 Author
Author  Correspondence author
Correspondence author
Triticeae Genomics and Genetics, 2024, Vol. 15, No. 5 doi: 10.5376/tgg.2024.15.0023
Received: 10 Aug., 2024 Accepted: 16 Sep., 2024 Published: 20 Sep., 2024
Wang Y.L., and He Z.H., 2024, Polyploidy in Triticeae: genetic mechanisms and agronomic implications, Triticeae Genomics and Genetics, 15(5): 244-254 (doi: 10.5376/tgg.2024.15.0023)
Polyploidy plays a crucial role in crops of the Triticeae tribe (such as wheat, barley, and rye), significantly influencing their evolutionary history and agronomic traits. Through mechanisms such as genome duplication, gene rearrangement, and functional diversity, polyploidy has driven the adaptability and productivity of Triticeae crops. Understanding the genetic mechanisms of polyploidy is essential for improving these important crops. This study explores the genetic mechanisms underlying the formation of polyploidy in Triticeae and its impact on agronomic traits. By analyzing post-polyploidization genomic changes, epigenetic modifications, and gene expression regulation, this research reveals how polyploidy promotes the improvement and enhanced adaptability of Triticeae crops. Additionally, it summarizes the applications of polyploidy in modern breeding and discusses its potential role in crop breeding and climate change adaptation in the future. Polyploidy not only has profound effects on the evolution of Triticeae crops but also provides important genetic resources for crop improvement. By gaining a deeper understanding of the molecular basis of polyploidy, breeders can leverage its advantages to enhance crop yield, disease resistance, and drought tolerance. Furthermore, polyploidy holds great significance in addressing complex genetic patterns and optimizing breeding strategies, contributing to the solutions for the challenges faced by modern agriculture.
1 Introduction
Polyploidy, the condition of possessing more than two complete sets of chromosomes, is a significant evolutionary force in the plant kingdom. The Triticeae tribe, which includes economically important crops such as wheat, barley, and rye, is particularly rich in polyploid species. This phenomenon has played a crucial role in the evolutionary history and adaptation of these species, contributing to their genetic diversity and agronomic traits.
Polyploidy has been a recurring theme in the evolutionary history of plants, including the Triticeae tribe. The process of polyploidization involves whole-genome duplication, which can occur through autopolyploidy (duplication within a single species) or allopolyploidy (combining genomes from different species) (Huang and Zhu, 2018). In the Triticeae tribe, allopolyploidy is particularly prevalent and has been a major driver of speciation and adaptation (Jauhar, 2007). For instance, the evolutionary history of wheat involves multiple polyploidization events, leading to the formation of hexaploid bread wheat (Triticum aestivum) from its diploid and tetraploid ancestors (Middleton et al., 2014). These events have resulted in complex genomic architectures and have facilitated the adaptation of Triticeae species to diverse environments (Blasio et al., 2022).
Polyploidy has had profound agronomic implications for crops like wheat and barley. The duplication of entire genomes has led to increased genetic variation, which is a valuable resource for breeding and crop improvement (Renny-Byfield and Wendel, 2014). In wheat, polyploidy has contributed to traits such as increased grain size, improved stress tolerance, and higher yield potential (Jauhar, 2007). Similarly, barley has benefited from polyploidization through enhanced adaptability and resilience to environmental stresses (Middleton et al., 2014). The genomic complexity introduced by polyploidy also poses challenges, such as difficulties in genome assembly and analysis, but advances in sequencing technologies are helping to overcome these obstacles (Renny-Byfield and Wendel, 2014).
This study provides a comprehensive overview of the genetic mechanisms and agronomic impacts of polyploidy in the Triticeae tribe. By exploring the evolutionary processes that have shaped the genomes of polyploid Triticeae species, with a focus on wheat and barley, it discusses the effects of polyploidy on the agronomic traits of these crops and the potential for future crop improvement. Polyploidy has played a key role in the evolution and domestication of Triticeae crops. Its role in generating genetic diversity and enhancing agronomic traits highlights the importance of continued research in this field. By synthesizing current research findings, the study examines the genetic complexity and practical significance of polyploidy, aiming to deepen the understanding of polyploidy in Triticeae and its importance for agriculture and plant breeding, offering valuable insights for researchers and breeders.
2 Mechanisms of Polyploidy Formation in Triticeae
2.1 Types of polyploidy: autopolyploidy and allopolyploidy
Polyploidy, the condition of having more than two complete sets of chromosomes, is a significant evolutionary mechanism in plants, including the Triticeae tribe. There are two primary types of polyploidy: autopolyploidy and allopolyploidy. Autopolyploidy arises from the duplication of a single species' genome, leading to multiple sets of homologous chromosomes. This type of polyploidy can result in increased genetic material, which may provide a fitness advantage under certain environmental conditions (Svačina et al., 2020; Luque et al., 2022). On the other hand, allopolyploidy results from hybridization between two distinct species followed by chromosome doubling. This process combines divergent genomes, which can lead to novel genetic combinations and potentially new species (Huang and Zhu, 2018; Svačina et al., 2020).
The formation of autopolyploids and allopolyploids involves different genetic and cytological mechanisms. Autopolyploids often face challenges such as multivalent formation during meiosis, which can lead to aneuploid gametes and reduced fertility (Svačina et al., 2020). In contrast, allopolyploids must establish compatibility between the divergent genomes and their regulatory networks, which can result in rapid genomic and phenotypic changes (Chen, 2007; Blasio et al., 2022). Despite these challenges, both types of polyploidy have been crucial in the evolution and diversification of the Triticeae tribe, contributing to their adaptability and speciation.
2.2 Molecular mechanisms of genome duplication
The molecular mechanisms underlying genome duplication in polyploids involve a complex interplay of genetic and epigenetic factors. In autopolyploids, genome duplication typically occurs through errors in meiosis or mitosis, leading to the formation of unreduced gametes. These errors can be due to altered spindle organization, disturbed kinetochore function, or abnormal cytokinesis (Blasio et al., 2022). In allopolyploids, hybridization between different species is followed by chromosome doubling, which can be facilitated by similar meiotic errors or by somatic cell fusion (Mason and Wendel, 2020; Blasio et al., 2022).
Once genome duplication occurs, polyploids undergo significant genomic and transcriptomic changes. These changes include alterations in DNA sequence, chromatin modifications, and RNA-mediated pathways, which can affect gene expression and phenotypic variation (Chen, 2007). Homoeologous recombination, where related chromosomes from different subgenomes pair and exchange genetic material, is a common feature in newly formed allopolyploids. This process can lead to genomic instability but also provides opportunities for evolutionary novelty and adaptation (Mason and Wendel, 2020). The stabilization of polyploid genomes involves the suppression of homoeologous recombination and the establishment of new regulatory networks, which are critical for the successful establishment and persistence of polyploid species (Mason and Wendel, 2020; Blasio et al., 2022).
2.3 Timeline of polyploidy events in Triticeae evolution
The evolutionary history of the Triticeae tribe is marked by multiple polyploidy events, which have played a crucial role in their diversification and adaptation. Polyploidy events in Triticeae can be traced back to ancient whole-genome duplications, which have been followed by more recent polyploidization events. These events have contributed to the complex genomic architecture observed in modern Triticeae species (Figure 1) (Huang and Zhu, 2018; Peer et al., 2020). For instance, wheat (Triticum spp.) is a well-known allopolyploid that has undergone several rounds of hybridization and genome duplication, resulting in its current hexaploid form (Svačina et al., 2020; Blasio et al., 2022).
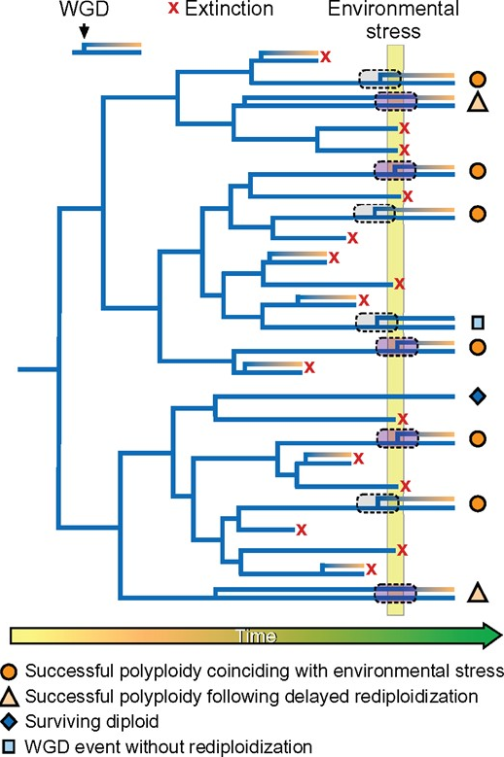 Figure 1 Whole genome duplication (WGD) events and their non-random association with environmental stresses in Triticeae evolution (Adapted from Peer et al., 2020) Image caption: The figure illustrates WGD events and their association with environmental stresses. Red crosses indicate extinction events, orange circles represent successful polyploidization events aligned with environmental stresses, light orange triangles signify delayed polyploidization followed by successful polyploidization, and dark blue diamonds indicate diploids that survived under environmental pressures. Gray squares mark WGD events that coincide with periods of global changes or extinction events. Branch color changes signify genome differentiation and rediploidization processes following chromosome doubling (Adapted from Peer et al., 2020) |
The timeline of polyploidy events in Triticeae evolution reveals a pattern of recurrent polyploidization, often associated with periods of environmental change or stress. These events have provided the genetic material necessary for adaptation to new environments and the development of new traits (Peer et al., 2020; Tossi et al., 2022). The repeated occurrence of polyploidy in Triticeae suggests that it is a key driver of their evolutionary success, enabling them to exploit diverse ecological niches and respond to changing environmental conditions (Huang and Zhu, 2018; Peer et al., 2020). Understanding the timeline and mechanisms of polyploidy in Triticeae can provide insights into the evolutionary processes that have shaped this important group of plants.
3 Genomic Changes Associated with Polyploidy in Triticeae
3.1 Chromosomal rearrangements and genomic shock post-polyploidization
Chromosomal rearrangements are a common consequence of polyploidization in Triticeae, driven by both internal factors such as nucleocytoplasmic interactions and external environmental influences. These rearrangements play a crucial role in the initial formation, stabilization, and establishment of polyploids (Zhu et al., 2024). For instance, hybrid breeding between common wheat and related wild species has led to significant chromosomal changes, such as the well-known 1BL/1RS translocation between wheat and rye, which has been widely utilized in wheat breeding programs (Wang et al., 2014). Additionally, studies on synthetic Arabidopsis allopolyploids have shown that genomic remodeling, including the activation of transposons and chromosomal fragment formation, can occur post-polyploidization, contributing to phenotypic instability and reduced fertility (Madlung et al., 2004).
Environmental factors also significantly influence chromosomal rearrangements in polyploid Triticeae species. Research on Kengyilia thoroldiana has demonstrated that populations from different environments exhibit varying frequencies of chromosome translocations, with higher rates observed in cold alpine and grassland environments compared to valley and lake-basin habitats (Wang et al., 2012). This suggests that environmental stress can exacerbate genomic shock, leading to increased chromosomal rearrangements and potentially driving the evolution of new ecotypes.
3.2 Gene duplication and its contribution to functional diversity in Triticeae
Gene duplication is a fundamental outcome of polyploidization, providing raw material for evolutionary innovation and functional diversification. In Triticeae, gene duplication resulting from polyploidization can lead to the development of novel gene functions and increased genetic diversity. For example, the process of homoeologous recombination in allopolyploids can generate new gene combinations and phenotypes, although it may also destabilize the karyotype and reduce fertility (Gaeta and Pires, 2010). This recombination-driven genetic variation is crucial for the adaptation and evolution of polyploid species.
Moreover, polyploidization-induced gene duplication can result in subfunctionalization, where duplicated genes diverge and specialize in different functions. This phenomenon has been observed in various plant polyploids, including Triticeae, where dynamic changes in gene expression and genomic organization occur post-polyploidization (Chen, 2007). These changes can lead to the emergence of new traits and improved adaptability, contributing to the success of polyploid species in diverse environments.
3.3 Epigenetic modifications and polyploid stability in Triticeae genomes
Epigenetic modifications play a vital role in maintaining genome stability and regulating gene expression in polyploid Triticeae. Polyploidization often triggers epigenetic changes, such as DNA methylation and histone modifications, which can influence gene expression and phenotypic variation. For instance, studies on synthetic plant allopolyploids have documented extensive changes in cytosine methylation and chromatin modifications, which are associated with gene repression, novel activation, and transposon activity (Chen and Ni, 2006). These epigenetic alterations help establish a compatible relationship between the divergent genomes in allopolyploids, facilitating their stabilization and evolution.
Furthermore, epigenetic mechanisms are crucial for the long-term stability of polyploid genomes. Research on natural allotetraploid Brachypodium hybridum has shown that while immediate genomic shock may not always occur, gradual epigenetic changes over evolutionary time contribute to genome diploidization and stability (Scarlett et al., 2022). These findings highlight the importance of epigenetic modifications in the adaptive evolution and domestication of polyploid Triticeae species, enabling them to thrive in various environmental conditions (Chen, 2007).
4 Polyploidy and Agronomic Trait Improvement in Triticeae
4.1 Polyploidy-driven enhancement of yield, drought tolerance, and disease resistance in Triticeae
Polyploidy has been shown to significantly enhance various agronomic traits in Triticeae, including yield, drought tolerance, and disease resistance. The induction of polyploidy can lead to increased biomass yield and improved stress tolerance. For instance, polyploid plants often exhibit enhanced tolerance to abiotic and biotic stresses, which can positively impact plant growth and net production (Tossi et al., 2022). Additionally, polyploidy can result in larger plant organs and increased cell size, which may contribute to higher yield potential (Corneillie et al., 2018). The genetic and physiological changes induced by polyploidy, such as increased gene expression and genome reorganization, are crucial for these improvements (Renny-Byfield and Wendel, 2014).
4.2 Genetic and phenotypic consequences of polyploidy on plant height, flowering time, and grain quality
Polyploidy can lead to significant genetic and phenotypic changes in Triticeae, affecting traits such as plant height, flowering time, and grain quality. Polyploid plants often exhibit delayed flowering and increased plant height due to changes in cell size and number (Corneillie et al., 2018). These changes can be attributed to the increased DNA content and altered gene expression patterns in polyploids (Balao et al., 2011). Moreover, polyploidy can influence grain quality by modifying the composition of bioactive compounds and altering metabolic pathways (Tavan et al., 2021). For example, polyploidy has been associated with changes in cell wall composition and sugar content, which can affect grain quality and processing characteristics (Corneillie et al., 2018).
4.3 Breeding strategies leveraging polyploid traits for crop improvement
Breeding strategies that leverage polyploid traits can significantly enhance crop improvement efforts in Triticeae. The use of synthetic polyploids and grafted crops can provide specific advantages, such as improved stress tolerance and yield. Breeding programs can exploit the genetic diversity and novel traits introduced by polyploidy to develop superior cultivars. For instance, the induction of polyploidy in rootstocks can enhance adaptation to biotic and abiotic stresses, while maintaining high yield and quality in the scion (Ruiz et al., 2022). Additionally, modern technologies, such as next-generation sequencing, can facilitate the identification and manipulation of polyploid traits, enabling more efficient breeding strategies (Renny-Byfield and Wendel, 2014). The integration of polyploidy into breeding programs holds great potential for the development of resilient and high-yielding Triticeae crops.
5 Gene Expression Modulation in Polyploid Triticeae
5.1 Subgenome interaction and differential gene expression in polyploid wheat and barley
Polyploidy in Triticeae, particularly in wheat and barley, results in complex interactions between subgenomes that significantly influence gene expression. In hexaploid wheat, the presence of three subgenomes (A, B, and D) leads to nonbalanced expression patterns in approximately 30% of homoeologous gene triads, with one homoeolog often showing higher or lower expression relative to the others. This differential expression is associated with epigenetic changes such as DNA methylation and histone modifications, and is influenced by the presence of transposable elements in gene promoters. Additionally, the transcriptional landscape of polyploid wheat reveals that homoeologous genes exhibit tissue-specific expression patterns, which are crucial for the plant's development and stress responses (Ramírez-González et al., 2018). The interaction between subgenomes also manifests in the form of gene dosage effects, where the deletion or addition of chromosomes in nullisomic-tetrasomic stocks leads to genome-wide changes in gene expression, affecting traits such as plant height and kernel number (Zhang et al., 2019).
5.2 Role of non-homoeologous recombination in modulating agronomically important traits
Non-homoeologous recombination plays a pivotal role in the genetic innovation and adaptation of polyploid Triticeae. In wheat, illegitimate recombination between homoeologous genes can lead to gene conversion events, which contribute to genome instability but also drive functional and structural innovation. This process affects a significant number of genes, including those involved in starch biosynthesis, thereby influencing important agronomic traits (Figure 2) (Liu et al., 2020). Moreover, the structural organization of chromosomes, including truncations and rearrangements, can modulate recombination frequencies, particularly in crossover-poor regions, enhancing the transfer of beneficial traits into crops (Naranjo, 2019). These recombination events are crucial for the introgression of useful genes and the overall improvement of crop performance.
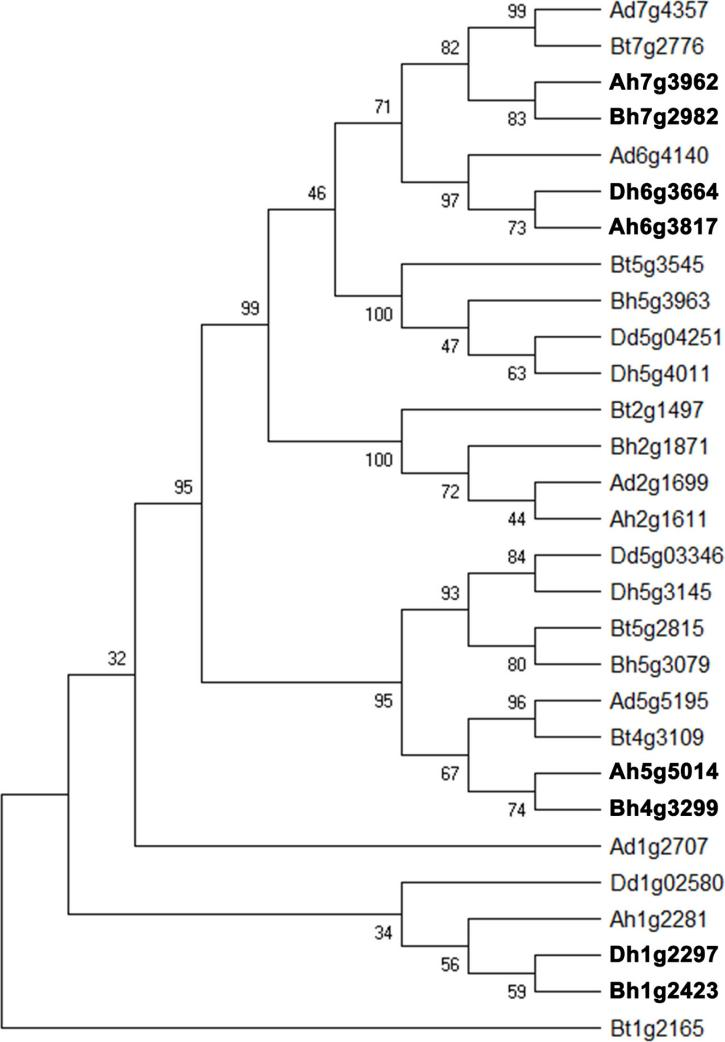 Figure 2 Phylogenetic tree of homoeologous gene quadruplicates involved in starch biosynthesis in Triticeae genomes (Adapted from Liu et al., 2020) Image caption: The figure displays a phylogenetic tree of homoeologous genes associated with starch biosynthesis, marked by different letters representing multiple subgenomes. Bt denotes the B subgenome of tetraploid wild wheat, Dd represents Aegilops tauschii, Ad stands for Triticum urartu, while Ah, Bh, and Dh represent the A, B, and D subgenomes of hexaploid bread wheat, respectively. Gene names in bold indicate genes that are regulated or have undergone functional changes in the starch synthesis process (Adapted from Liu et al., 2020) |
Liu et al. (2020) revealed the evolutionary branches of homoeologous genes in Triticeae species and the conservation and diversity of genes across different subgenomes during polyploid evolution. Non-homologous recombination plays a critical role in regulating key agronomic traits, such as starch biosynthesis, in crops like wheat. This genetic diversity allows for functional complementation between different subgenomes, giving polyploid plants an advantage in adaptability and growth performance. This study provides an important theoretical foundation for understanding the genetic improvement of essential crops like wheat, aiding in the enhancement of crop yield and quality through molecular breeding methods.
5.3 How polyploidy influences regulatory networks, transcription factors, and stress response genes in Triticeae
Polyploidy significantly impacts the regulatory networks, transcription factors, and stress response genes in Triticeae. The presence of multiple subgenomes in polyploid wheat leads to complex regulatory interactions that are essential for the plant's adaptation and stress responses. For instance, the transcriptional landscape of polyploid wheat during embryogenesis and grain development shows that gene expression is shaped by the contributions of the A, B, and D subgenomes, with each subgenome playing distinct roles in different developmental stages (Xiang et al., 2019). Additionally, histone modifications, such as H3K4me3 and H3K27me3, are conserved across subgenomes and are crucial for regulating gene expression during domestication and ploidy transitions (Lv et al., 2021). These epigenetic modifications help maintain genome stability and ensure proper gene function under various environmental conditions. Furthermore, the coordination of homoeologous gene expression in response to stress is facilitated by coexpression networks, which reveal extensive interactions between genes throughout the plant's development (Liu et al., 2020). This intricate regulatory framework enables polyploid Triticeae to exhibit enhanced resilience and adaptability, making them valuable for agricultural improvement.
By understanding these genetic mechanisms, researchers and breeders can develop strategies to manipulate gene expression and improve agronomic traits in polyploid Triticeae, ultimately enhancing crop performance and resilience.
6 Applications of Polyploidy in Triticeae Crop Breeding
6.1 Case studies of successful breeding improvements through polyploidy
Polyploidy has been instrumental in the breeding of various Triticeae crops, leading to significant improvements in yield, quality, and stress resistance. For instance, synthetic polyploids have been used to enhance specific traits in crop varieties, such as environmental adaptation and yield, by modifying certain plant phenotypes while maintaining fundamental characteristics (Ruiz et al., 2020). In wheat, the use of hexaploid varieties has facilitated the identification of homoeologous relationships between chromosomes, aiding in the transfer of valuable agronomic traits from related species (Naranjo, 2019). Additionally, polyploidy has been employed to overcome the non-viability and infertility of interspecific hybrids, resulting in the development of seedless polyploid cultivars and increased resistance to biotic and abiotic factors (Trojak-Goluch et al., 2021).
6.2 Technical challenges and solutions in incorporating polyploidy into breeding programs
Incorporating polyploidy into breeding programs presents several technical challenges, including the complexities of genome assembly and the potential for undesired phenotypic effects. The genome-wide analysis of polyploid crops has historically lagged behind that of diploid crops due to difficulties in genome assembly, which arise from the combination of evolutionarily diverged genomes into a single nucleus and the significant size of polyploid genomes (Renny-Byfield and Wendel, 2014). However, advancements in next-generation sequencing and other molecular tools have begun to address these challenges, enabling better understanding and manipulation of polyploid genomes (Renny-Byfield and Wendel, 2014; Kyriakidou et al., 2018). Additionally, the use of antimitotic agents such as colchicine, oryzalin, and trifluralin has been effective in inducing polyploidy, although the efficiency of genome duplication can vary based on species, cultivar, genotype, and tissue type (Trojak-Goluch et al., 2021).
6.3 Future potential of polyploid crops in breeding programs
The future potential of polyploid crops in breeding programs is vast, particularly in the context of climate change and the need for crops that can withstand biotic and abiotic stresses. Polyploidy offers the potential for increased allelic diversity, heterozygosity, and novel phenotypic variation, which are crucial for crop improvement (Udall and Wendel, 2006). The generation of synthetic polyploids as a breeding strategy has already led to the development of new and improved cultivars, and ongoing research aims to further explore the mechanisms underlying polyploidy-induced novelty (Iannicelli et al., 2020). As our understanding of polyploid genomes continues to grow, particularly through the use of modern genomic technologies, the ability to harness the benefits of polyploidy for crop breeding will likely expand, offering new opportunities for the development of resilient and high-yielding Triticeae crops (Bharadwaj, 2015; Heslop-Harrison et al., 2022).
7 Challenges in Managing Polyploidy in Triticeae Breeding Programs
7.1 Difficulties in mapping complex traits in polyploid genomes
Mapping complex traits in polyploid genomes presents significant challenges due to the intricate nature of polyploid inheritance and the presence of multiple homologous chromosomes. Polyploid organisms, such as those in the Triticeae tribe, often exhibit complex genetic interactions that complicate the identification and mapping of quantitative trait loci (QTL). The presence of multiple gene copies and structural variations, such as homeologous exchanges, further complicates genetic mapping efforts (Bourke et al., 2018; Schiessl et al., 2019). Advanced genomic tools and techniques, including genome-wide association studies (GWAS) and quantitative trait analysis, are essential to overcome these challenges and facilitate the accurate mapping of complex traits in polyploid Triticeae species (Bourke et al., 2018).
7.2 Breeding barriers due to complex inheritance patterns in polyploid species
Polyploid species exhibit complex inheritance patterns that pose significant barriers to breeding programs. The presence of multiple sets of chromosomes can lead to genome instabilities, chromosome imbalances, and regulatory incompatibilities, which in turn affect reproductive success and fertility (Comai, 2005; Chen, 2007). Additionally, the intricate interactions between redundant genes and the potential for non-Mendelian inheritance patterns further complicate breeding efforts (Wendel, 2004). These challenges necessitate the development of specialized breeding strategies and the use of advanced genomic tools to manage the complex inheritance patterns in polyploid Triticeae species (Chen, 2007; Bourke et al., 2018).
7.3 Overcoming linkage drag and sterility issues in polyploid triticeae crops
Linkage drag and sterility are significant issues in polyploid Triticeae crops that hinder breeding progress. Linkage drag occurs when undesirable traits are co-inherited with beneficial traits due to their close proximity on the chromosome, making it difficult to separate them through traditional breeding methods (Schiessl et al., 2019). Sterility issues arise from the complex interactions between divergent genomes in allopolyploids, leading to reproductive failures and reduced fertility (Chen, 2007; Suissa et al., 2021). Overcoming these challenges requires innovative breeding techniques, such as the use of genomic selection and the development of synthetic polyploids, to break linkage drag and enhance fertility in polyploid Triticeae crops (Chen, 2007; Bourke et al., 2018; Schiessl et al., 2019).
8 Future Research Directions
8.1 Further exploration of the long-term effects of polyploidy on Triticeae gene function
Polyploidy has been a significant evolutionary force in plants, leading to gene function innovation and species diversification. However, the long-term effects of polyploidy on gene function in Triticeae remain underexplored. Future research should focus on understanding how polyploidy-induced gene fractionation and sub-/neo-functionalization impact the genetic and phenotypic traits of Triticeae over extended periods. This could involve longitudinal studies on polyploid Triticeae species to monitor changes in gene expression, genome stability, and trait development under various environmental conditions (Zhang et al., 2019; Heslop-Harrison et al., 2022).
8.2 Optimization of molecular breeding techniques for polyploid crops
The complexity of polyploid genomes poses challenges for traditional breeding methods. Advances in next-generation sequencing and genome editing technologies offer new opportunities for optimizing molecular breeding techniques for polyploid crops. Future research should aim to develop high-throughput genotyping and phenotyping platforms tailored for polyploid species, as well as refine CRISPR/Cas9 and other genome editing tools to efficiently target multiple gene copies. This will facilitate the rapid development of polyploid crops with improved traits such as yield, stress tolerance, and nutritional value (Kyriakidou et al., 2018; Pourkheirandish et al., 2020; Huang and Li, 2024).
8.3 Role of polyploid crops in addressing global climate change and food security
Polyploid crops have the potential to play a crucial role in mitigating the impacts of climate change and ensuring food security. Research should focus on identifying and harnessing the genetic diversity within polyploid species to develop crops that are resilient to biotic and abiotic stresses. This includes exploring the adaptive potential of polyploid crops through gene flow and introgression from wild relatives, as well as investigating the role of polyploidy in enhancing stress tolerance and biomass production under changing environmental conditions (Ruiz et al., 2020; Lovell et al., 2021; Cheng et al., 2022; Tossi et al., 2022).
9 Concluding Remarks
Polyploidy has played a fundamental role in the evolution and agricultural production of the Triticeae tribe, which includes key cereal crops such as wheat, barley, and rye. The process of polyploidization, involving whole-genome duplication, has been a significant evolutionary force, driving the diversification and adaptation of plant species. In Triticeae, polyploidy has facilitated the development of new genetic variants and the introgression of beneficial traits from wild relatives, enhancing crop resilience and productivity. The ability of polyploid species to undergo rapid genomic changes and adapt to various environmental conditions underscores their importance in both natural ecosystems and agricultural settings.
Polyploidy significantly impacts genetic diversity, adaptability, and crop breeding. The duplication of entire genomes results in genetic redundancy, which can lead to novel gene functions and increased genetic variation. This genetic diversity is crucial for the adaptability of polyploid species, allowing them to thrive in diverse and changing environments. In crop breeding, polyploidy has been exploited to enhance desirable traits such as disease resistance, yield, and stress tolerance. For instance, hexaploid wheat has benefited from the introgression of genes from wild relatives, improving its agronomic performance. The ability to manipulate polyploid genomes through modern breeding techniques and next-generation sequencing has further expanded the potential for crop improvement.
The future application and research directions of polyploidy in agriculture are promising. Advances in genomic technologies, such as genome sequencing and editing, provide new opportunities to explore and harness the benefits of polyploidy for crop improvement. Research should focus on understanding the mechanisms underlying polyploidy-induced phenotypic changes and their implications for crop performance. Additionally, the development of synthetic polyploids and their use in grafted crops could offer innovative solutions for enhancing crop resilience to biotic and abiotic stresses, particularly in the context of climate change. Continued exploration of the genetic and epigenetic changes associated with polyploidy will be essential for optimizing the use of polyploid crops in sustainable agriculture.
Acknowledgments
We thank Dr. Feng for critically reading the manuscript and providing constructive feedback.
Conflict of Interest Disclosure
The authors affirm that this research was conducted without any commercial or financial relationships that could be construed as a potential conflict of interest.
Balao F., Herrera J., and Talavera S., 2011, Phenotypic consequences of polyploidy and genome size at the microevolutionary scale: a multivariate morphological approach, The New phytologist, 192(1): 256-265.
https://doi.org/10.1111/j.1469-8137.2011.03787.x
Bharadwaj D.N., 2015, Polyploidy in crop improvement and evolution, In: Bahadur B., Venkat Rajam M., Sahijram L., Krishnamurthy K., (eds) Plant Biology and Biotechnology, Springer, New Delhi, pp.619-638.
https://doi.org/10.1007/978-81-322-2286-6_24
Blasio F., Prieto P., Pradillo M., and Naranjo T., 2022, Genomic and meiotic changes accompanying polyploidization, Plants, 11(1): 125.
https://doi.org/10.3390/plants11010125
Bourke P., Voorrips R., Visser R., and Maliepaard C., 2018, Tools for genetic studies in experimental populations of polyploids, Frontiers in Plant Science, 9: 513.
https://doi.org/10.3389/fpls.2018.00513
Chen Z., 2007, Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids, Annual Review of Plant Biology, 58: 377-406.
https://doi.org/10.1146/annurev.arplant.58.032806.103835
Chen Z., and Ni Z., 2006, Mechanisms of genomic rearrangements and gene expression changes in plant polyploids., BioEssays, 28(3): 240-252.
https://doi.org/10.1002/bies.20374
Cheng A., Hanafiah N., Harikrishna J., Eem L., Baisakh N., and Mispan M., 2022, A reappraisal of polyploidy events in grasses (poaceae) in a rapidly changing world, Biology, 11(5): 636.
https://doi.org/10.3390/biology11050636
Comai L., 2005, The advantages and disadvantages of being polyploid, Nature Reviews Genetics, 6: 836-846.
https://doi.org/10.1038/nrg1711
Corneillie S., Storme N., Acker R., Fangel J., Bruyne M., Rycke R., Geelen D., Willats W., Vanholme B., and Boerjan W., 2018, Polyploidy affects plant growth and alters cell wall composition1, Plant Physiology, 179: 74-87.
https://doi.org/10.1104/pp.18.00967
Gaeta R., and Pires J., 2010, Homoeologous recombination in allopolyploids: the polyploid ratchet, The New Phytologist, 186(1): 18-28.
https://doi.org/10.1111/j.1469-8137.2009.03089.x
Heslop-Harrison J., Schwarzacher T., and Liu Q., 2022, Polyploidy: its consequences and enabling role in plant diversification and evolution, Annals of Botany, 131: 1-10.
https://doi.org/10.1093/aob/mcac132
Huang G., and ZhuY., 2018, Plant polyploidy and evolution, Journal of Integrative Plant Biology, 61(1): 4-6.
https://doi.org/10.1111/jipb.12758
Huang Y.M., and Li J.Q., 2024, Functional and structural insights from the Oryza genome: implications for crop enhancement, Rice Genomics and Genetics, 15(4): 164-176.
https://doi.org/10.5376/rgg.2024.15.0017
Iannicelli J., Guariniello J., Tossi V., Regalado J., Ciaccio L., Baren C., Álvarez S., and Escandon A., 2020, The “polyploid effect” in the breeding of aromatic and medicinal species, Scientia Horticulturae, 260: 108854.
https://doi.org/10.1016/j.scienta.2019.108854
Jauhar P., 2007, Meiotic restitution in wheat polyhaploids (amphihaploids): a potent evolutionary force, The Journal of Heredity, 98(2): 188-193.
https://doi.org/10.1093/jhered/esm011
Kyriakidou M., Tai H., Anglin N., Ellis D., and Strömvik M., 2018, Current strategies of polyploid plant genome sequence assembly, Frontiers in Plant Science, 9: 1660.
https://doi.org/10.3389/fpls.2018.01660
Liu C., Wang J., Sun P., Yu J., Meng F., Zhang Z., Guo H., Wei C., Li X., Shen S., and Wang X., 2020, Illegitimate recombination between homeologous genes in wheat genome, Frontiers in Plant Science, 11: 1076.
https://doi.org/10.3389/fpls.2020.01076
Lovell J., MacQueen A., Mamidi S., Bonnette J., Jenkins J., Napier J., Sreedasyam A., Healey A., Session A., Shu S., Barry K., Bonos S., Boston L., Daum C., Deshpande S., Ewing A., Grabowski P., Haque T., Harrison M., Jiang J., Kudrna D., Lipzen A., Pendergast T., Plott C., Qi P., Saski C., Shakirov E., Sims D., Sharma M., Sharma R., Stewart A., Singan V., Tang Y., Thibivillier S., Webber J., Weng X., Williams M., Wu G., Yoshinaga Y., Zane M., Zhang L., Zhang J., Behrman K., Boe A., Fay P., Fritschi F., Jastrow J., Lloyd-Reilley J., Martínez-Reyna J., Matamala R., Mitchell R., Rouquette F., Ronald P., Saha M., Tobias C., Udvardi M., Wing R., Wu Y., Bartley L., Casler M., Devos K., Lowry D., Rokhsar D., Grimwood J., Juenger T., and Schmutz J., 2021, Genomic mechanisms of climate adaptation in polyploid bioenergy switchgrass, Nature, 590: 438-444.
https://doi.org/10.1038/s41586-020-03127-1
Luque J., Moreno E., Kovalsky I., Seijo J., and Neffa V., 2022, Polyploidy, genome size variation and diversification in an autopolyploid complex: the case of Turnera sidoides (Passifloraceae, Turneroideae), Systematics and Biodiversity, 20: 1-18.
https://doi.org/10.1080/14772000.2022.2036854
Lv Z., Li Z., Wang M., Zhao F., Zhang W., Li C., Gong L., Zhang Y., Mason A., and Liu B., 2021, Conservation and trans-regulation of histone modification in the A and B subgenomes of polyploid wheat during domestication and ploidy transition, BMC Biology, 19: 1-16.
https://doi.org/10.1186/s12915-021-00985-7
Madlung A., Tyagi A., Watson B., Jiang H., Kagochi T., Doerge R., Martienssen R., and Comai L., 2004, Genomic changes in synthetic Arabidopsis polyploids, The Plant Journal : for Cell and Molecular Biology, 41(2): 221-230.
https://doi.org/10.1111/j.1365-313X.2004.02297.x
Mason A., and Wendel J., 2020, Homoeologous exchanges, segmental allopolyploidy, and polyploid genome evolution, Frontiers in Genetics, 11: 1014.
https://doi.org/10.3389/fgene.2020.01014
Middleton C., Senerchia N., Stein N., Akhunov E., Keller B., Wicker T., and Kilian B., 2014, Sequencing of chloroplast genomes from wheat, barley, rye and their relatives provides a detailed insight into the evolution of the Triticeae tribe, PLoS ONE, 9(3): e85761.
https://doi.org/10.1371/journal.pone.0085761
Naranjo T., 2019, The Effect of chromosome structure upon meiotic homologous and homoeologous recombinations in Triticeae, Agronomy, 9(9): 552.
https://doi.org/10.3390/agronomy9090552
Peer Y., Ashman T., Soltis P., and Soltis D., 2020, Polyploidy: an evolutionary and ecological force in stressful times, The Plant Cell, 33: 11-26.
https://doi.org/10.1093/plcell/koaa015
Pourkheirandish M., Golicz A., Bhalla P., and Singh M., 2020, Global role of crop genomics in the face of climate change, Frontiers in Plant Science, 11: 922.
https://doi.org/10.3389/fpls.2020.00922
Ramírez-González R., Borrill P., Lang D., Harrington S., Brinton J., Venturini L., Davey M., Jacobs J., Ex F., Pasha A., Khedikar Y., Robinson S., Cory A., Florio T., Concia L., Juery C., Schoonbeek H., Steuernagel B., Xiang D., Ridout C., Chalhoub B., Mayer K., Benhamed M., Latrasse D., Bendahmane A., Wulff B., Appels R., Tiwari V., Datla R., Choulet F., Pozniak C., Provart N., Sharpe A., Paux E., Spannagl M., Bräutigam A., and Uauy C., 2018, The transcriptional landscape of polyploid wheat, Science, 361(6403): eaar6089.
https://doi.org/10.1126/science.aar6089
Renny-Byfield S., and Wendel J., 2014, Doubling down on genomes: polyploidy and crop plants, American Journal of Botany, 101(10): 1711-1725.
https://doi.org/10.3732/ajb.1400119
Ruiz M., Oustric J., Santini J., and Morillon R., 2020, Synthetic polyploidy in grafted crops, Frontiers in Plant Science, 11: 540894.
https://doi.org/10.3389/fpls.2020.540894
Scarlett V., Lovell J., Shao M., Phillips J., Shu S., Lusinska J., Goodstein D., Jenkins J., Grimwood J., Barry K., Chalhoub B., Schmutz J., Hasterok R., Catalán P., and Vogel J., 2022, Multiple origins, one evolutionary trajectory: gradual evolution characterizes distinct lineages of allotetraploid Brachypodium, Genetics, 223(2): iyac146.
https://doi.org/10.1093/genetics/iyac146
Schiessl S., Katche E., Ihien E., Chawla H., and Mason A., 2019, The role of genomic structural variation in the genetic improvement of polyploid crops, The Crop Journal, 7(2): 127-140.
https://doi.org/10.1016/j.cj.2018.07.006
Suissa J., Kinosian S., Schafran P., Bolin J., Taylor W., and Zimmer E., 2021, Homoploid hybrids, allopolyploids, and high ploidy levels characterize the evolutionary history of a western North American quillwort complex (Isoëtes), Molecular Phylogenetics and Evolution, 166: 107332.
https://doi.org/10.1016/j.ympev.2021.107332
Svačina R., Sourdille P., Kopecký D., and Bartoš J., 2020, Chromosome pairing in polyploid grasses, Frontiers in Plant Science, 11: 10.
https://doi.org/10.3389/fpls.2020.01056
Tavan M., Sarikhani H., Mirjalili M., Rigano M., and Azizi A., 2021, Triterpenic and phenolic acids production changed in Salvia officinalis via in vitro and in vivo polyploidization: a consequence of altered genes expression, Phytochemistry, 189: 112803.
https://doi.org/10.1016/j.phytochem.2021.112803
Tossi V., Tosar L., Laino L., Iannicelli J., Regalado J., Escandon A., Baroli I., Causin H., and Pitta-Álvarez S., 2022, Impact of polyploidy on plant tolerance to abiotic and biotic stresses, Frontiers in Plant Science, 13: 869423.
https://doi.org/10.3389/fpls.2022.869423
Trojak-Goluch A., Kawka-Lipińska M., Wielgusz K., and Praczyk M., 2021, Polyploidy in industrial crops: applications and perspectives in plant breeding, Agronomy, 11(12): 2574.
https://doi.org/10.3390/agronomy11122574
Udall J., and Wendel J., 2006, Polyploidy and crop improvement, Crop Science, 46: S-3-S-14.
https://doi.org/10.2135/cropsci2006.07.0489tpg
Wang Q., Gao A., Yang X., and Li L., 2014, Chromosome changes after polyploidization in Triticeae, Journal of Systematics and Evolution, 52(6): 790-793.
https://doi.org/10.1111/jse.12125
Wang Q., Liu H., Gao A., Yang X., Liu W., Li X., and Li L., 2012, Intergenomic rearrangements after polyploidization of Kengyilia thoroldiana (Poaceae: Triticeae) affected by environmental factors, PLoS ONE, 7(2): e31033.
https://doi.org/10.1371/journal.pone.0031033
Wendel J., 2004, Genome evolution in polyploids, Plant Molecular Biology, 42: 225-249.
https://doi.org/10.1023/A:1006392424384
Xiang D., Quilichini T., Liu Z., Gao P., Pan Y., Li Q., Nilsen K., Venglat P., Esteban E., Pasha A., Wang Y., Wen R., Zhang Z., Hao Z., Wang E., Wei Y., Cuthbert R., Kochian L., Sharpe A., Provart N., Weijers D., Gillmor S., Pozniak C., and Datla R., 2019, The transcriptional landscape of polyploid wheats and their diploid ancestors during embryogenesis and grain development, Plant Cell, 31: 2888-2911.
https://doi.org/10.1105/tpc.19.00397
Zhang K., Wang X., and Cheng F., 2019, Plant polyploidy: origin, evolution, and its influence on crop domestication, Horticultural Plant Journal, 5(6): 231-239.
https://doi.org/10.1016/j.hpj.2019.11.003
Zhang R., Geng S., Qin Z., Tang Z., Liu C., Liu D., Song G., Li Y., Zhang S., Li W., Gao J., Han X., and Li G., 2019, The genome-wide transcriptional consequences of the nullisomic-tetrasomic stocks for homoeologous group 7 in bread wheat, BMC Genomics, 20: 1-17.
https://doi.org/10.1186/s12864-018-5421-3
Zhu Q., Zhang X.L., Zhang H., Li J., Wang C.L., Lee D.S., and Chen L.J., 2024, Autotetraploid rice hybrids: overcoming sterility barriers for enhanced heterosis, Molecular Plant Breeding, 15(4): 167-177.
https://doi.org/10.5376/mpb.2024.15.0017
.png)
. PDF(550KB)
. FPDF(win)
. FPDF(mac)
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Yali Wang
. Zhonghui He
Related articles
. Triticeae
. Polyploidy
. Genetic mechanisms
. Gene expression
. Agronomic traits
Tools
. Email to a friend
. Post a comment


